How to use NeuroJSON.io
NeuroJSON.io website provides an intuitive interface for users of any experience level to easily navigate across potentially vast amount of public/free datasets curated by NeuroJSON developers and contributors. It offers web-based interfaces to preview the data items before download, and provides convenient search functionalities to help users identify the desired datasets for their analyses.
Before we start, please always keep in mind that NeuroJSON.io contains many databases; each database contains many datasets; each dataset contains one or many subjects.
- 1. Navigate databases in a browser
- 1.1. Landing page
- 1.2. Database information
- 1.3. Browse and query a database
- 1.4. Preview a volume object in a dataset
- 1.5. Preview a mesh object
- 3. Advanced search
- 4. Download a database and read file locally
- 5. Access data using RESTful APIs
- 6. Dynamically downloading/caching linked binary data resources
1. Navigate databases in a browser
1.1. Landing page
The NeuroJSON.io main page serves as the entry point for exploring NeuroJSON free datasets, referred to as the "data constellation", showing a self-organizing map of existing data collections in the form of a 3-D force graph (thanks to the open-source 3d-force-graph Javascript library). Key features of this graph include
- each node on the graph is a "data collection", containing many datasets
- the size of each node is correlated to the number of datasets contained
- the color of each node is computed based on the included data modalities (MRI, EEG, MEG, fNIRS etc)
- links are added between nodes containing the same modality, bundling them together; the color of the link is modality specific
- use your cursor, you can drag, rotate, zoom in/out of the graph; clicking on any of the nodes, you could see the detailed metadata of the data collection in a side bar
1.2. Database information
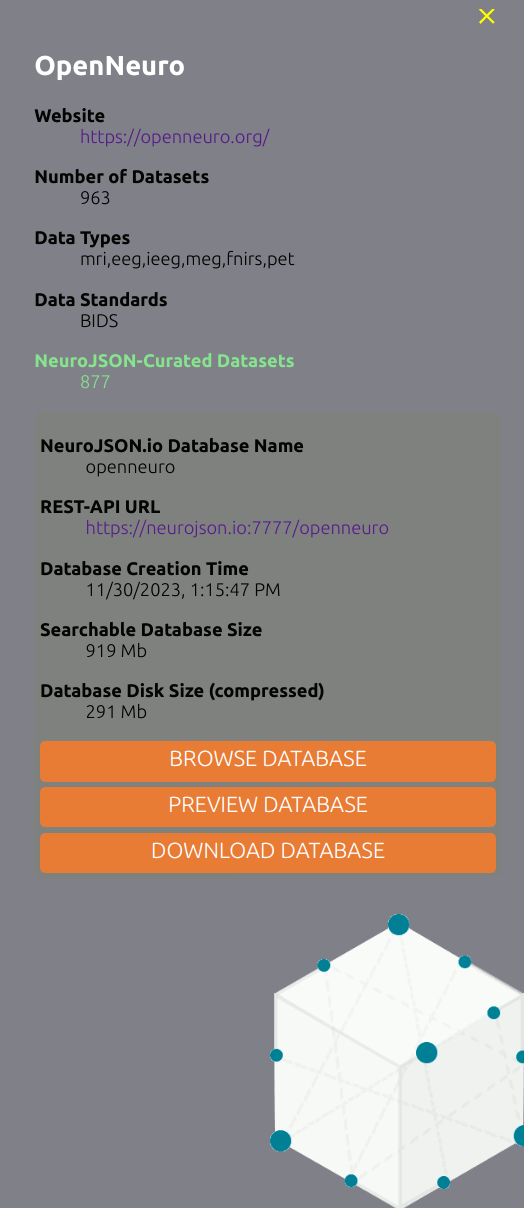
A side panel is shown on the left whenever a user select a database (a node) on the "data constellation". In this panel, additional details regarding the data collection are listed, including the upstream website, data modality types, standards etc. Specifically, if such data collection has been curated by NeuroJSON, an additional block is shown on the
1.3. Browse and query a database
When clicking on the "Browse Database" button in the database information panel, your browser will load a webpage that help you navigate and search different datasets within the selected database.
If a database is conforming to the BIDS specification, such as openneuro, a summary table for each containing datasets will be shown. By default, 25 databases is shown on a page; one can click on the page numbers on the top to navigate between different pages to browse the summary for each dataset.
Video demo
1.4. Preview a volume object in a dataset
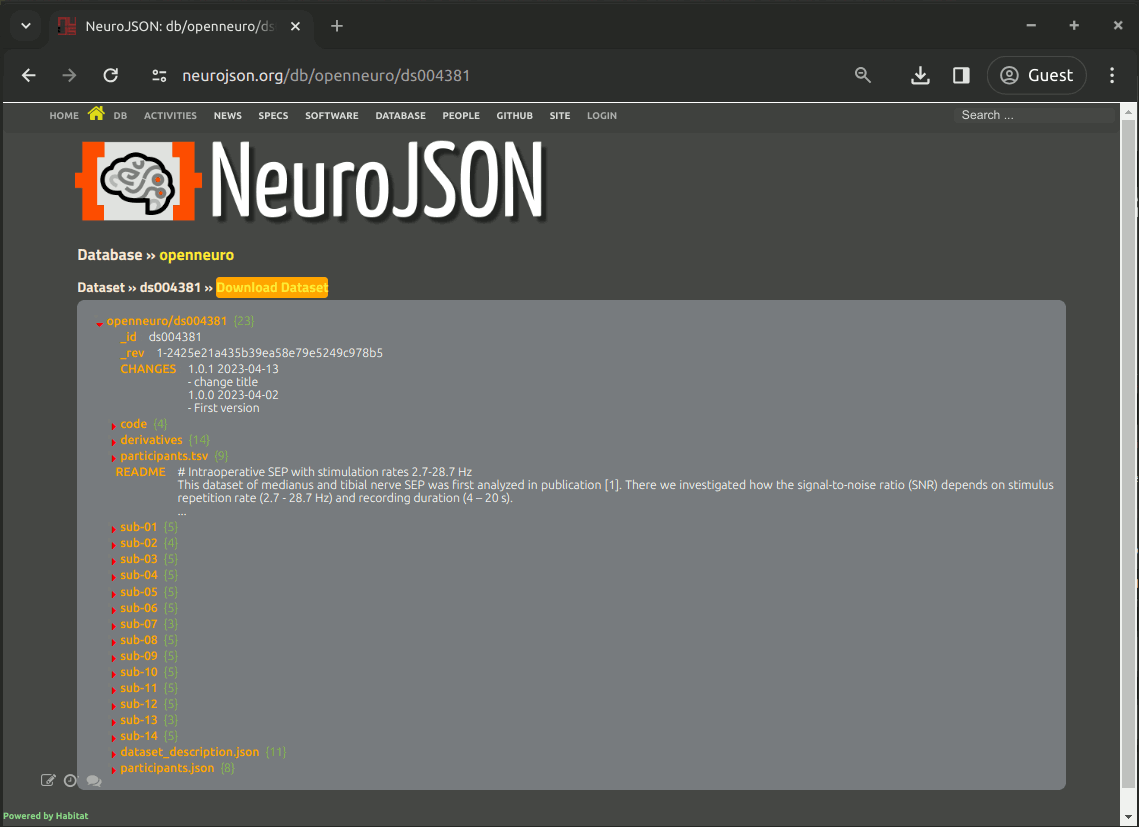
When selecting a dataset from a database, a page, in the URL format of https://neurojson.org/db/dbname/dsname, is shown to display the entire searchable content of the full dataset in a tree-like viewer. You can think of the tree as a folder structure, with each item representing a data file or a subfolder. As a matter of fact, that's how we convert BIDS datasets to the JSON format.
By expanding each items in the JSON tree, the content or sub-folder is shown.
Video demo
1.5. Preview a mesh object
We use JSON to encode diverse types of neuroimaging data structures, including our JMesh specification for describing shapes, tetrahedral and triangular meshes. Here is an example showing how to use the preview feature to render a surface mesh object embedded inside the dataset.
Video demo
2. Search datasets and subject metadata
By taking advantage of the automated metadata extraction and aggregation functions provided in the CouchDB/modern NoSQL databases, we can search across large amount of datasets with the lightweight automatically indexed metadata.
A user can perform efficient search using the below interface
a video tutorial on how to use this function is provided below
Video demo
3. Advanced search
CouchDB supports fine-grained search of a full database (including all datasets contained) using various APIs. The `_find` API is one of such powerful data search tools.
A _find API queries a dataset by posing a JSON object describing the search criteria. It contains a "selector" object specifying the conditions and a "fields" object specifying the returned fields. The detailed documentation of the _find API can be found here
In NeuroJSON.io, we provide a _find API query builder for each database. In the database information, one simply select the "Search Database" button, or type https://neurojson.io/find/<databasename>; as the URL.
Below is a video demo on how to use the query building to filter datasets.
Video demo
4. Download a database and read file locally
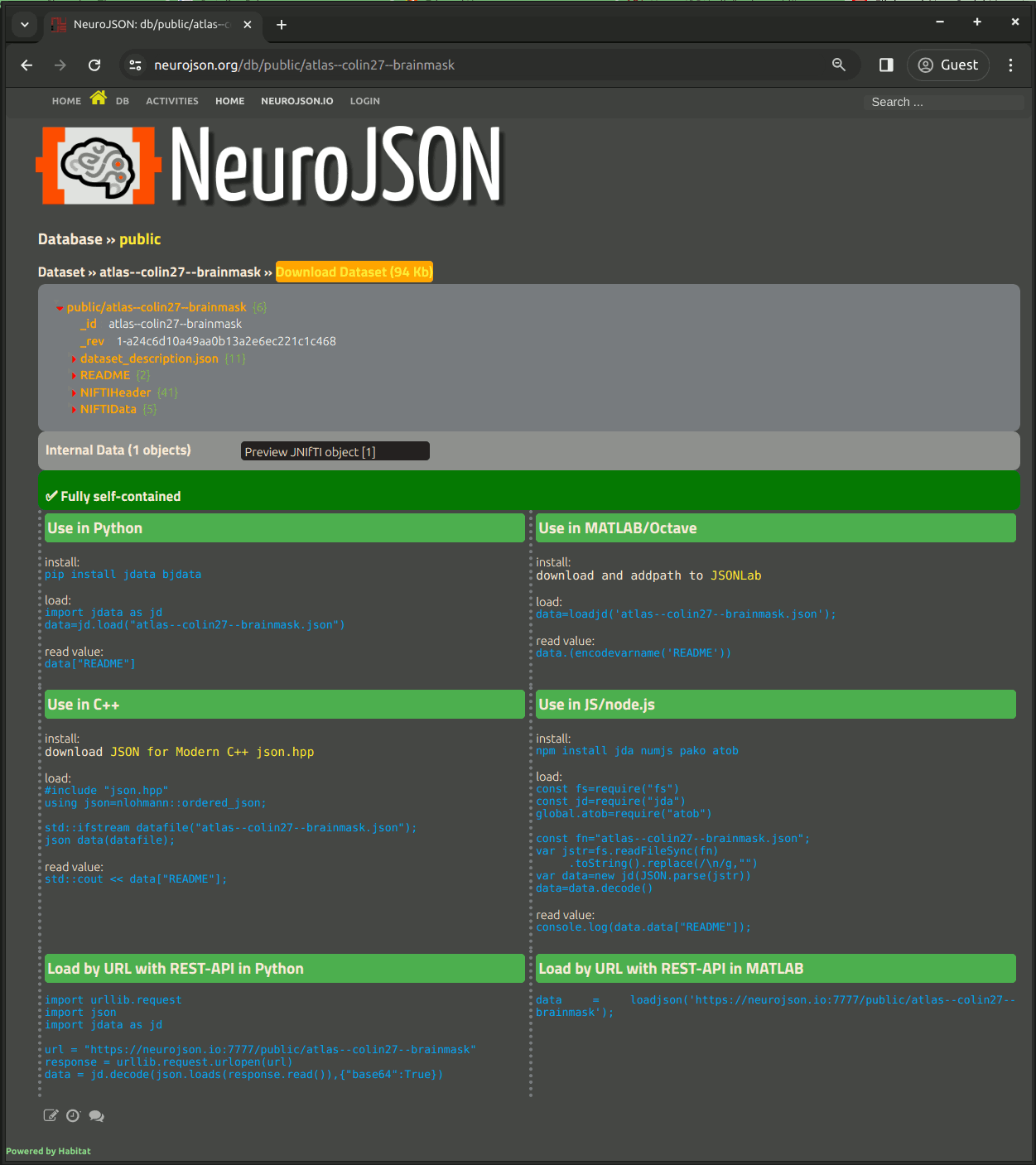
On every dataset page, right above the JSON tree, one can see a "Download" button. Clicking on this button downloads the JSON-encoded dataset to your local drive.
At the bottom of the page, we provide command lines for each programming environments how to load/decode the NeuroJSON-curated JSON based dataset files.
5. Access data using RESTful APIs
One can access all datasets from all public databases hosted on NeuroJSON.io using the CouchDB supported RESTful APIs. Because RESTful APIs are URL-based, all the data resources on NeuroJSON.io can be virtually accessed from any programming environments, including web and cloud applications.
Here are the RESTful API formats to access the data on NeuroJSON.io
| URL | Returned results | Examples |
|---|---|---|
GET https://neurojson.io:7777/sys/registry | list all public databases hosted on NeuroJSON.io and their metadata | example |
GET https://neurojson.io:7777/dbname | list the information related to a database named "dbname" | example |
GET https://neurojson.io:7777/dbname/_all_docs | list all the documents stored within the database named "dbname" | example |
GET https://neurojson.io:7777/dbname/_all_docs?limit=X&skip=Y | list all documents in database "dbname", list the first X docs starting from the Y-th | example |
GET https://neurojson.io:7777/dbname/dsname | list the full content of the dataset "dsname" in database "dbname" | example |
GET https://neurojson.io:7777/dbname/dsname?revs=true | list the full content of dataset "dsname" in "dbname" with revision list | example |
GET https://neurojson.io:7777/dbname/dsname?rev=revhash | retrieve the specific revision "revhash" of document "dsname" in "dbname" | example |
POST https://neurojson.io:7777/dbname/_find | use the CouchDB _find API to query specific data fields | see below |
| Data query example: When calling the _find API to query a database, a user post a JSON construct, like below, that defines a set of query criteria; Here, the "selector" field asks CouchDB to find participant age below 20; the "fields" item asks CouchDB to return the specified data items once the records are found; the "limit" field sets a maximum length for each returned query. | ||
Video demo
6. Dynamically downloading/caching linked binary data resources
A large dataset typically contains several gigabytes to hundreds of gigabytes of binary data files. Conventionally, a user has to download the entire zipped data package (many GBs in size) regardless whether they need all the embedded large data files or not.
Using our lightweight REST-API and JSON-encoded datasets, a user can selectively download and locally cache those data files that are only necessary to the analysis, making it much more efficient to start analyzing large datasets. Here are 3 tutorials showing how to do this in MATLAB and Octave.
Tutorial Part 1 - Use REST-API for data download and query a dataset
Tutorial Part 2 - Downloading and rendering linked data files
Tutorial Part 3 - Using JSON API on Octave
![[Home]](upload/neurojson_banner_plain.png)