Why NeuroJSON
NeuroJSON.io serves human-readable, searchable neuroimaging datasets using universally accessible JSON format and URL-based RESTful APIs. NeuroJSON.io is built upon highly scalable document-store NoSQL database technologies, specifically, open-source Apache CouchDB engine, that can handle millions of datasets without major performance penalties. It provides fine-grained data search capabilities to allow users to find, preview and re-combine complex data records from public datasets before download.
- 1. What problems are we addressing?
- 2. What solutions does NeuroJSON provide?
- 3. NeuroJSON guiding principles
- 4. NeuroJSON data-exchange platform architecture
- 5. Mapping of neuroimaging datasets to a CouchDB database
1. What problems are we addressing?
Traditional neuroimaging data sharing has been largely focused on file-based data sharing that faces challenges in 1) handling diverse and complex file formats involved, 2) lack of standardization in naming conventions, and 3) lack of human-readable and machine-actionable metadata. The emerging BIDS (brain imaging data structure) standard has greatly simplified and homogenized file-package based dataset organization, making sure that the data files and folders are organized in simple, consistent and meaningful semantic order, with restricted file types, accompanied with human/machine-readable metadata, both in the modality data-file level and the dataset level.
However, file-package based data sharing still faces a number of key challenges
- without searchability and findability, it is difficult to scale towards exponentially growing public neuroimaging datasets, both in size and complexity
- many of the data files, even with restrictions under the BIDS specifications, are in binary form and are not directly human readable or searchable; their utilities are dependent on the continual maintenance of file parsers; future discontinuity or upgrade of certain file formats may jeopardize the long-term viability of the dataset
- increasing use of complex data analysis pipelines via automated and distributed cloud-computing services demands more flexible and fine-grained data access; disseminating large zipped packages could add significant overhead to data processing and maintenance.
2. What solutions does NeuroJSON provide?
The NIH funded NeuroJSON project addresses the needs for scalability and long-term viability of scientific datasets by adopting the JSON format and NoSQL database technologies that have been extensively developed and widely adopted by the IT industries over the last few decades.
JSON is a human-readable hierarchical data format that is extensively used among web and native applications for ubiquitous data exchange. It is an internationally standardized format and has a large tool ecosystem, filled with numerous free parsers and utilities developed for nearly every programming language and environment in existence. JSON is supported by default within many prominent programming languages such as Python, Perl, MATLAB/Octave, and JavaScript, with lightweight parsers available for most other programming environments.
To enable rapid search and manipulation of massive amount of complex data, a new kind of database engine, NoSQL database, has been developed and broadly used in routine handling of large data produced by online and cloud-based applications. Different from traditional table-based relational databases, NoSQL databases can effectively handle and manipulate hierarchical data records and JSON is often used as the native data exchange format for many NoSQL database engines.
The NeuroJSON project first define a set of lightweight specifications to "wrap" common neuroimaging data files into a JSON constructs. These specifications ranges from JData specification -- responsible for mapping common scientific data structures such as tables and N-D arrays to JSON structures, to JNIfTI specification -- responsible for mapping a NIfTI data file to a JSON construct, to JSNIRF specification -- responsible for mapping an HDF5 based SNIRF data files to JSON, and JMesh specification -- responsible for portable exchange of discrete shape/mesh data, among others.
Based on these specifications, we have developed a set of converters that can convert common neuroimaging data files, including the folder structure defined by BIDS specification, and extract all searchable metadata and content to JSON based files. With these JSON encoded files, we can "upload" the searchable portion of the dataset, potentially those from many many datasets, to a highly scalable NoSQL database to facilitate fast and complex data search.
At NeuroJSON.io, we run an instance of Apache CouchDB server to host NeuroJSON curated datasets. We chose CouchDB because it is fully open-source (compared to MongoDB), and supports automatic synchronization between multiple database instances.
3. NeuroJSON guiding principles
The long-term vision of the NeuroJSON project and the software ecosystem were built upon the below guiding principles:
- 1. Open-data MUST BE provided in "source-code" data format
2. Open-data MUST BE human-understandable - It has been widely understood that open-source software must ensure "Freedom 1" - that the software must be understood by users. It ensures such freedom by demanding the access to the source code (which must be human-understandable). Although the FAIR principle has been widely recognized as the guiding principle for sharing open-data, it has not been recognized that open-data must be provided in the "source-code" format to ensure that such data can be fully understood and reused by future users. It is our vision that JSON and our JSON-based JData data annotations was designed fill this need and serve as the universal "source-code format" for all open-data sharing. Adopting and demanding the provision of such free-data source format is the only way to ensure they can be reused in the future.
- 3. Diverse data formats do harm to open-science and open-data
- Diverse data formats, often developed in ad hoc fashion by software developers or targeted at application-specific needs, are liability towards the long-term viability of the data they store. Most data formats are not self-contained. Their utility relies on the proper implementation of separate parser libraries and specifications that are external to the data files. When the parser upgrades or retires, or the external specification advances to new versions, data stored in the legacy formats are rendered unusable unless the parsers for older format are continue to be maintained, which adds costs and complexity. Scientific community should consider adopting standardized container formats to store value data assets without relying on application-specific data formats.
- 4. Small is beautiful
- This is one of the well recognized tenets of "Unix Philosophy". Same philosophy should be applied to open-data sharing. Conventional data sharing via large-sized all-in-one zipped file packages fails to enable users to read, search, understand and select the data in the package unless they fully download the package. This is extremely inefficient and not scalable. Extracting human-readable small-sized metadata from complex datasets is the key to enable rapid, efficient access and offer highly efficient data manipulation, inteoperation and scalable data processing.
- 5. Metadata is your data!
- In the US legal system (as well as most of the world), raw measurement data are considered "facts", and thus are not copyrightable (therefore, experimenters are NOT in a position to set any license to the data because they do not own the copyright). However, metadata and annotations that experimenters use to organize their measurement datasets to make those understandable are, in many cases but not all, copyrightable. In a way, the metadata/annotation portion of a dataset is the only part that the experimenters can "own" and decide how to disseminate. Creating a data dissemination system that specifically focuses on indexing, distributing and handling human-understandable metadata not only makes the data dissemination extremely compact and efficient (see "Small is beautiful" principle above), it also readily enables scalability (that easily grows to large datasets) and findability/searchable that the FAIR principle demands.
4. NeuroJSON data-exchange platform architecture
To systematically address scalability and searchability in scientific data sharing, we have built a versatile data exchange platform and address challenges at multiple levels. The diagram below shows the overall design architecture. Let us walk you through our platform architecture from the bottom-level to the top-level step-by-step.
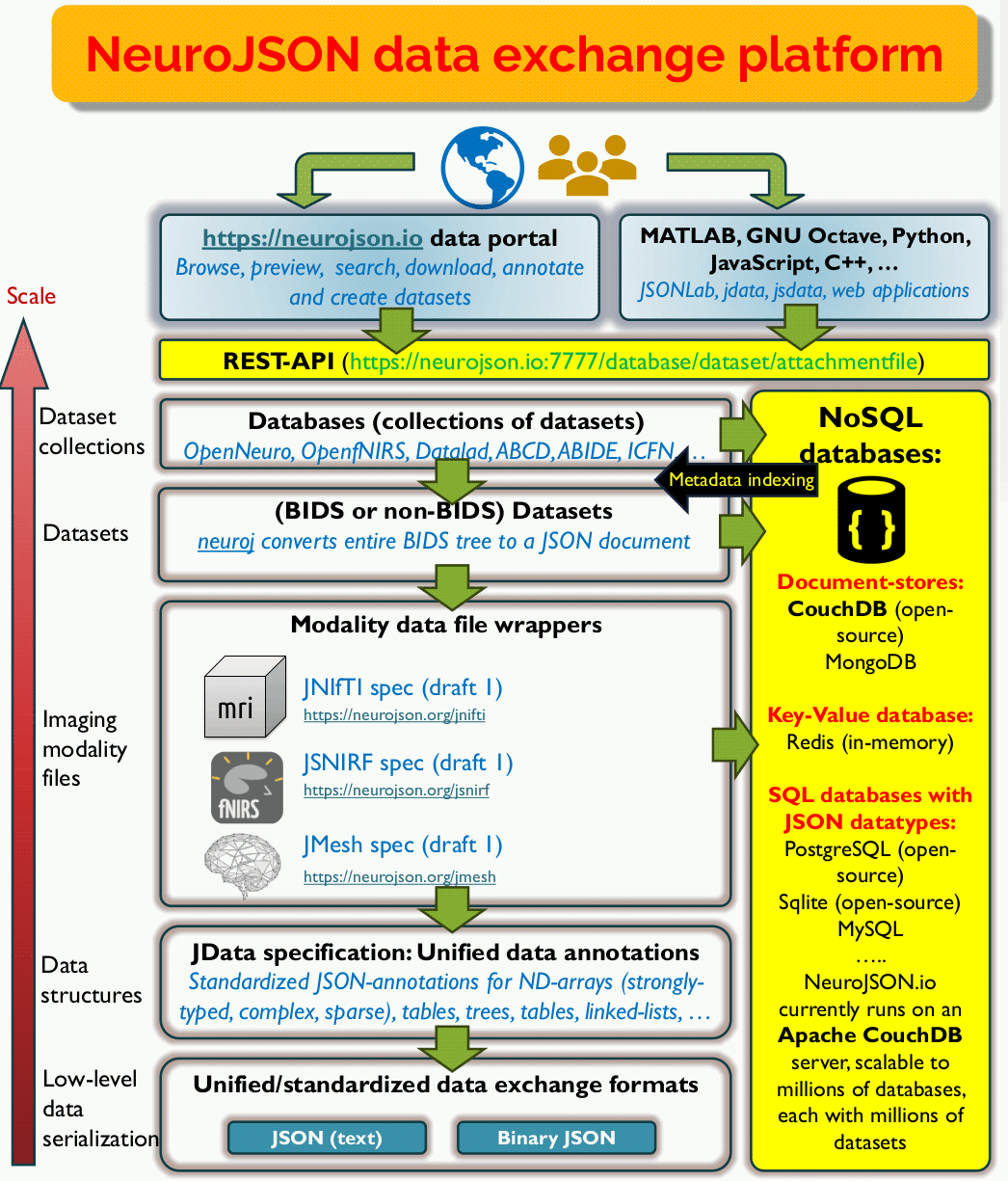
At the lowest level (data serialization), the foundation of the NeuroJSON platform is solidly seated in the JSON format - an internationally standardized, portable, ubiquitous and human-readable format that can be used in any programming language and understood by any user, now and future. It is JSON's universal presence and future-proof human-readability that had won our bet to carry valuable scientific data resources, including both raw data and metadata, for generations to come. The vast ecosystem of tools, including numerous free JSON parsers, standards such as JSON Path, JSON reference, JSON schema, JSON-LD, many NoSQL databases and much more, are readily at our disposal once our data are JSON-compliant.
We also recognize that there is a need for efficiency. For that purpose, we supplement JSON with an interchangeable binary JSON interface, Binary JData or BJData format for applications where high IO speed and smaller disk space is desired. This is a new format derived from a widely-used binary JSON format called Universal Binary JSON (UBJSON). Compared to other more sophisticated binary JSON variants such as MessagePack, CBOR, BSON etc, UBJSON's KISS (keep it simple and stupid) design philosophy has the closest mirror to the core spirit of JSON. We made some key modifications to UBJSON to specially optimize it towards carrying scientific/neuroimaging data, including native support for N-D strongly-typed arrays. In the past years, our C++ BJData parsers has been included in JSON for Modern C++, a highly popular C++ JSON library that has received over 4 millions of downloads. Our BJData format support were also included in Python, MATLAB/Octave, JavaScript and C. JSON and binary JSON can be losslessly converted from one to the other.
After building the low-level serialization format interface, we subsequently moved to standardizations of scientific data structures. This leads to our JData specification - a lightweight semantic layer that is aimed to describe all common scientific data structures using pure JSON annotations. We intentionally implemented JData completely with JSON annotation/keywords, making all NeuroJSON data files 100% JSON compatible without introducing customized syntax. Examples of the JData annotated data structures include N-D arrays, typed/complex/sparse N-D arrays, tables, trees, graphs, linked lists, binary streams, etc. Binary N-D array data can be losslessly enclosed by a JSON construct with optional data compression (supporting multiple codecs, including the high-performance blosc2 meta-compressor). Ultra-lightweight JData annotation encoders/decoders have been made widely available for MATLAB/Octave, Python, JavaScript etc.
With our standardized/portable data stream and data structures at the foundation, we can readily approach neuroimaging data sharing (or any general scientific data sharing at large) and "modernize" them to become scalable, searchable and interoperable.
At the smallest scale, a neuroimaging dataset consists of many data files that enclose different pieces of information regarding the diverse imaging modalities and procedures involved. We built a series of lossless JSON wrappers to map these data files to a JSON "container" or "wrapper". These file-level JSON wrappers include the JNIfTI spec (JSON-wrapper of the commonly used NIfTI-1/2 data files), JSNIRF spec (JSON-wrapper of the SNIRF format used in fNIRS), JMesh spec (for storing mesh, geometry and shape data in a JSON form). Emerging neuroimaging data sharing standards, such as BIDS and Zarr, already started using JSON sidecar files as the main carriers for metadata; these JSON files are readily usable in NeuroJSON's ecosystem without needing a wrapper or converter.
At the middle-level, neuroimaging data are typically shared in the unit of "datasets". It has been a challenging task of sharing datasets that are made of diverse measurements and modalities that involve many data files. Inconsistent naming conventions, file/folder organization schemes, and non-standard metadata names and storage locations have set major barriers for effective imaging data sharing over the past. Fortunately, community-driven efforts, especially [BIDS standard, have been greatly reducing this barrier and make it possible to parse/exchange data under a predictable, semantically meaningful, yet still simple folder/file structure. To enclose an entire dataset using our JSON framework, we have developed BIDS-to-JSON converters (such as neuroj or jbids) that can iterate through every data file (.nii.gz, .tsv, .json, .bval, etc) under a dataset, separating the searchable metadata with the non-searchable binary content, and map each part to their JSON equivalence, and then combine all searchable content to a single JSON "digest" file.
NeuroJSON project is aimed at an even more ambitious goal - we don't want to just stop at the file-package level for data sharing, but think bigger at the levels of collections of datasets (we call it a "database") or many collections of datasets (i.e. databases of databases) - how can we effectively represent, search, locate, interact, and process data on such a scale? Now, NoSQL database technologies enter the scene. NoSQL database is the umbrella term describing a new generation of database architectures that can handle complex, heterogeneous and hierarchical data as opposed to the table-like data structure supported in traditional relational databases. NoSQL databases have seen rapid growth over the last few decades and build the backbones for modern-day information technology and application data exchange. There are many flavors of NoSQL databases, including document-store databases (CouchDB, MongoDB), key-value-pair databases (Radis, Valkey), graph-databases (Neo4j), among others. Because JSON is hierarchical and heterogeneous in nature, unsurprisingly, it has been serving as the primary data exchange format for nearly all of these NoSQL database engines. As long as we can convert datasets and databases of neuroimaging data to JSON, we can readily store such data in the highly optimized NoSQL database engines and achieve scalable, rapid and versatile data search, manipulation and dissemination.
This leads to the top level of our architecture and our vision how neuroimaging datasets (or any scientific dataset) should be accessed and processed in the future. User should not just limit themselves to processing locally downloaded single imaging data files or single dataset, rather, they should use our highly scalable NoSQL database interfaces (commonly known as the RESTful-API, which provides a URL-based universal access to online/cloud content) to rapidly search, locate desired data files that match their needs across large collections of datasets (or mega-database), both interactively and programmably in automated pipelines. The exchanged data are all encoded in human/machine readable JSON packets that is lightweight, versatile, and language/platform neutral. The access and compute can be done inside local hardware or high-performance large-scale cloud based systems. The representation and storage of data in NeuroJSON framework are always human-understandable and searchable, making them easily reusable in the future.
5. Mapping of neuroimaging datasets to a CouchDB database
To carry neuroimaging datasets in a CouchDB database, we use the following mapping schemes to convert the logical structure of datasets/dataset collections to the hierarchies provided by a CouchDB (databases, documents, attachments etc)
| Data logical structure | CouchDB object | Examples |
|---|---|---|
| a dataset collection | a CouchDB database | openneuro, dandi, openfnirs,... |
| a dataset | a CouchDB document | ds000001, ds000002, ... |
| files and folders related to a subject | JSON keys inside a document | sub-01, sub-1/anat/scan.tsv,... |
| human-readable binary content (small) | an attachment to a document | .png, .jpg, .pdf, ... |
| non-searchable binary content (large) | _DataLink_ JSON key | "_DataLink_":"http://url/to/ds/filehash.jbd" |
![[Home]](upload/neurojson_banner_plain.png)